I wished to do some machine learning for binary classification. Binary classification is perhaps the most basic of all supervised learning problems. Unsurprisingly julia has many libraries for it. Today we are looking at: LIBLINEAR (linear SVMs), LIBSVM (Kernel SVM), XGBoost (Extreme Gradient Boosting), DecisionTrees (RandomForests), Flux (neural networks), TensorFlow (also neural networks).
In this post we are only concentrating on their ability to be used for binary classification. Most (all) of these do other things as well. We’ll also not really be going into exploring all their options (e.g. different types of kernals).
Furthermore, I’m not rigeriously tuning the hyperparameters so this can’t be considered a fair test for performance. I’m also not performing preprocessing (e.g. many classifies like it if you standarise your features to zero mean unit variance). You can look at this post more as talking above what code for that package looks like, and this is roughly how long it takes and how well it does out of the box.
It’s more of a showcase of what packages exist.
For TensorFlow and Flux, you could also treat this as a bit of a demo in how to use them to define binary classifiers.
Since they don’t do it out of the box.
This post, like most of my posts, is backed by a jupyter notebook.
Feel free, encouraged even, to download and run that, or view it on github, etc.
Also to raise issues on that repository.
The Task: Predict if that part of the Australian Flag is Blue
This is on the mildly gnarly side of binary classification problems. The classifying regions are:
- Not linearly seperable
- You can’t draw a line such that on one since is all the blue parts and on the other is all the nonblue parts.
- Not connected
- The stars, for example, are entirely separated by blue background regions
- Not convex
- With-in a section of one color, you can draw a line between two points in the same colored region and have it exit that section, then reenter.
- Unbalanced classes
- Most of the image is blue.
So it seams like a good, difficult, problem.
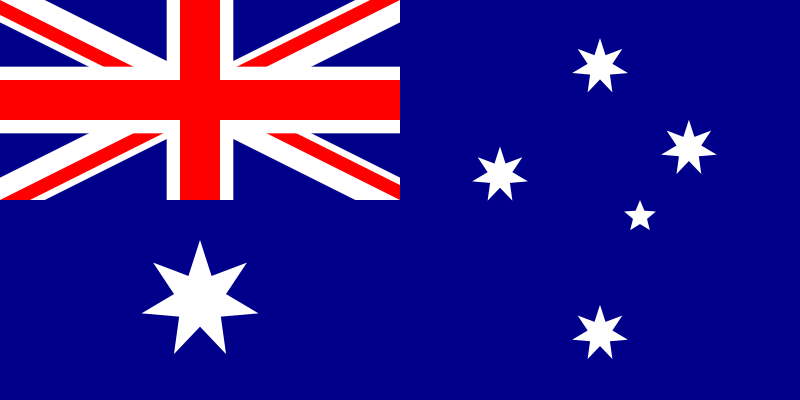
Data Generation
An image of the flag gives us one datum per pixel. We’re going to sample that, just so that plotting is easier.
Input:
using Images, FileIOInput:
img = load(download("https://upload.wikimedia.org/wikipedia/en/thumb/b/b9/Flag_of_Australia.svg/320px-Flag_of_Australia.svg.png"));Output:
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 3333 100 3333 0 0 2494 0 0:00:01 0:00:01 --:--:-- 2494Input:
isblue(pixel) = pixel.b > pixel.r && pixel.b > pixel.gOutput:
isblue (generic function with 1 method)
Input:
colorview(Gray, .!(isblue.(img)))Output:

Input:
const all_feature1 = Vector{Float64}()
const all_feature2 = Vector{Float64}()
const all_labels = Vector{Bool}()
@inbounds for ind in eachindex(IndexCartesian(), img)
pixel = img[ind]
push!(all_labels, isblue(pixel))
push!(all_feature1, ind.I[1])
push!(all_feature2, ind.I[2])
end
const all_features = [all_feature1'; all_feature2']
# standard julia Observations are in final index form (i.e columns of matrixes)
Any[all_features; all_labels']Output:
3×51200 Array{Any,2}:
1.0 2.0 3.0 4.0 … 157.0 158.0 159.0 160.0
1.0 1.0 1.0 1.0 320.0 320.0 320.0 320.0
false false false false true true true true
Normally I would do this data munging using MLDataUtils.jl, which I have blogged about before (though it might be nice to few more posts about it, it is a great package, and I don’t know that I’ve fully covered its capacities).
But since I am already about to introduce 6 packages, I thought I would minimize talking about other ones.
Input:
const all_inds = shuffle(1:length(all_labels))
const test_inds = all_inds[1:end÷5] # first 20%
const train_inds = all_inds[19end÷20:end] # last 5%
const test_features = all_features[:, test_inds]
const test_labels = all_labels[test_inds]
const train_features = all_features[:, train_inds]
const train_labels = all_labels[train_inds];Input:
using Plots
pyplot() # Using PyPlot, because the SVGs that GR makes kill browsers with too many paths at this scale
function plotflag(xs,ys; title="")
scatter(xs[2,:],-xs[1,:]; zcolor=ys,
markersize=2, markerstrokealpha=0, bg=colorant"gray", seriescolor=:blues, title=title)
endInput:
plotflag(train_features, train_labels, title="Training Data")Output:
Input:
plotflag(test_features, test_labels, title="Test Data")Output:
Interface
As was discussed on the julia slack yesterday. There is a real problem with a lack of consistency in our ML packages right now.
So I am going to take a leaf from XKCD #927, and define one.

StatsBase.fit(modeltype, features, labels)returns a model of that type that is trained on those features and labels.- Since we are only interested in binary classification, labels witll be an
AbstractVector{Bool}with one entry per column of the feature matrix
- Since we are only interested in binary classification, labels witll be an
StatsBase.fit!(model, features, labels)- Mutating form of the above. (
fitis basically a construtor) - useful for allowing retraining/hot-starting
- Mutating form of the above. (
StatsBase.predict(model, features)returns a vector of estimated probabilities of classification being true- one entry per column in features.
Something like this is actually in use in a bunch of places already, just not these packages, it seems..
Some packages (LibSVM, DecisionTrees.jl) use the same names, from ScikitLearnBase, but they go sideways (i.e. observations in rows, Python style).
I think the real solution to a good interface does need to be thinking more like (or using) MLDataUtils.jl, which is observation dimention agnostic, defaulting to normal julia practice (ObsDim.Last()).
Using these we can define our metrics, etc. It might be nicer to be using MLMetrics.jl to do this for us. But I’ll just do it simply here.
Input:
import StatsBase: fit!, fit, predict
classify(model, features) = predict(model, features).>0.5
accuracy(model, features, ground_truth_labels) = mean(classify(model, features) .== ground_truth_labels)Output:
accuracy (generic function with 1 method)
Evaluation function
Given a common interface we can write one function to evaluate them all.
Accessing our training and test data as a global variable.
(Obviously not a good idea normally).
Input:
percent(x) = @sprintf("%0.2f%%", 100*x)
function evaluate(modeltype)
@time model = fit(modeltype, train_features, train_labels)
println("$modeltype Train accuracy: ", percent(accuracy(model, train_features, train_labels)))
println("$modeltype Test accuracy: ", percent(accuracy(model, test_features, test_labels)))
#this is calculating the predict twice (since we did it to report accuaracy already), but predict is cheap
plotflag(test_features, predict(model, test_features); title=string(modeltype))
endOutput:
evaluate (generic function with 1 method)
LIBLINEAR.jl
The linear SVM. Possibly the weakest classifier in modern use. It actually works ok for a lot of higher dimentional problems. In high dimensions it is easier for things to be linearly seperable.
It surprises me that the C backend was only created in 2008.
R.-E. Fan, K.-W. Chang, C.-J. Hsieh, X.-R. Wang, and C.-J. Lin. LIBLINEAR: A Library for Large Linear Classification, Journal of Machine Learning Research 9(2008), 1871-1874. Software available at http://www.csie.ntu.edu.tw/~cjlin/liblinear
Because we are interesting in getting probabilities back from predict
we are restricted to using L2R_LR and L1R_LR solver types, which are logistric regression.
This could probably be relaxed for most applications (but might break those metrics defintions above).
Input:
using LIBLINEAR
function fit(::Type{LinearModel}, features, labels; solver_type=LIBLINEAR.L2R_LR, kwargs...)
linear_train(labels, features; solver_type=solver_type, kwargs...)
end
function predict(model::LinearModel, features)
classes, probs = linear_predict(model, features; probability_estimates=true)
vec(probs)
endOutput:
predict (generic function with 2 methods)
Input:
evaluate(LinearModel)Output:
0.735264 seconds (158.51 k allocations: 8.306 MiB)
LIBLINEAR.LinearModel Train accuracy: 79.85%
LIBLINEAR.LinearModel Test accuracy: 80.88%We can see from the plot that it is basically a gradient, of how much blue is in an area. This is as expected.
LIBSVM.jl
The more general SVM package. We’re here for its kernal SVM classifers. Again I am surprised that the backend was created so recently: 2005
Since version 2.8, it implements an SMO-type algorithm proposed in this paper: R.-E. Fan, P.-H. Chen, and C.-J. Lin. Working set selection using second order information for training SVM. Journal of Machine Learning Research 6, 1889-1918, 2005. https://www.csie.ntu.edu.tw/~cjlin/libsvm/
We’re looking at SVC, in this example.
The other types of interst here would be NuSVC, and LinearSVC (but we got that covered by LIBLINEAR)
Input:
import LIBSVM:svmtrain, SVM, svmpredict
function fit(::Type{SVM{Bool}}, features, labels; solver_type=LIBLINEAR.L2R_LR, kwargs...)
#could use ScikitLearnBase.fit!(SVC, features, Float64.(labels)), but it doesn't take extra args same way.
svmtrain(features, labels; probability=true, kwargs...)
end
function predict(model::SVM{Bool}, features)
classes, probs = svmpredict(model, features)
probs[1,:]
endOutput:
predict (generic function with 3 methods)
Input:
evaluate(SVM{Bool})Output:
5.028115 seconds (128.57 k allocations: 6.933 MiB)
LIBSVM.SVM{Bool} Train accuracy: 99.96%
LIBSVM.SVM{Bool} Test accuracy: 91.69%This is pretty nice. I guess the RBF keneral is not capturing enough area around the training points though, as we have a lot of blue mixed in with the white sections.
DecisionTree.jl
Shallow decision trees are one of the only ML systems, that are not black box. Anyone can look in and see how they work. And while, the human domain expert might not make those rules themselves, they can say that “yes those rules make sense”. This makes DecisionTrees way easier to “sell” to a consumer.
This package has pruned trees, random forests and adaptive-boosted decision stumps.
They are all useful; and all suitable. I’ve not evaluating the AdaBoostStumpClassifier as when I tried it threw errors.
I’m going to just wrap the ScikitLearnBase interface here, as DecisionTree.jl seems to be alwary sideway anyway.
Input:
using DecisionTree
import ScikitLearnBase
function fit!(model::ScikitLearnBase.BaseClassifier, features, labels)
#could use ScikitLearnBase.fit!(SVC, features, Float64.(labels)), but it doesn't take extra args same way.
ScikitLearnBase.fit!(model, features', labels)
end
function predict(model::ScikitLearnBase.BaseClassifier, features)
ScikitLearnBase.predict_proba(model, features')[:,2]
end
function fit(kind::Type{<:ScikitLearnBase.BaseClassifier}, features, labels, args...;kwargs...)
fit!(kind(args...;kwargs...), features, labels) #ScikitLearn stores most parameters in the model type
endOutput:
WARNING: using DecisionTree.fit! in module Main conflicts with an existing identifier.
WARNING: using DecisionTree.predict in module Main conflicts with an existing identifier.fit (generic function with 15 methods)
Input:
evaluate(DecisionTreeClassifier)Output:
2.299324 seconds (856.07 k allocations: 46.456 MiB, 1.36% gc time)
DecisionTree.DecisionTreeClassifier Train accuracy: 100.00%
DecisionTree.DecisionTreeClassifier Test accuracy: 95.52%Input:
evaluate(RandomForestClassifier)Output:
1.117075 seconds (411.63 k allocations: 40.382 MiB, 1.92% gc time)
DecisionTree.RandomForestClassifier Train accuracy: 99.53%
DecisionTree.RandomForestClassifier Test accuracy: 95.80%I think it is neat seeing the “blockyness”.
This is because the decisions at each node of the tree are axis aligned.
e.g. 25 < x && x < 30 && 140 < y && y < 160.
Note, also how the random forest has a lot more fractional probabilities. This is because it is combining the output of many decision trees.
XGBoost.jl
eXtreme Gradient Boosting.
These are boosting tree ensembles.
They are all about adding extra rules to just handling the errors existing rules don’t catch.
This basically makes training fast, and simply improves the results.
They win Kraggle all the time.
Right now, it hasn’t had a release tagged, so is broken on julia 0.6
This can be solved using Pkg.checkout("XGBoost"); Pkg.build("XGBoost").
Input:
import XGBoost: xgboost, BoosterInput:
function fit(::Type{Booster}, features, labels; num_rounds=16, eta=1, max_depth = 16, kwargs...)
xgboost(features', num_rounds; label=labels, objective = "binary:logistic", eta=eta, max_depth=max_depth, silent=true, kwargs...)
end
function predict(model::Booster, features)
XGBoost.predict(model, features')
endOutput:
predict (generic function with 5 methods)
I note that fit! could be defined to hot-start using the (method shown here)[https://github.com/dmlc/XGBoost.jl/blob/master/demo/boost_from_prediction.jl].
Input:
evaluate(Booster)Output:
0.977256 seconds (187.56 k allocations: 10.091 MiB)
XGBoost.Booster Train accuracy: 99.84%
XGBoost.Booster Test accuracy: 96.56%Notice it also has axis aligned decisions. This is I think the best performing model for this task. Though, if the hyper-parameters are tuned differently sometimes it loses out to random forests.
As an aside, I bet feature augmentation via rotational transforms works amazingly for decision trees and relatd methods (like XGBoost). Since they are axis aligned, adding an extra feature that is just the existing features rotated 45°, would allow for them to easier capture decision boundries that are at angles.
NaiveBayes.jl
Surprisingly, even though this is the most probabilistic model,
it is the most reluctant to give up class probabilities.
It’s also almost (but not quiet) using the interface I want to use.
It’s predict gives back a class assigment.
(It predict_proba might be better for what I am doing anyway, except NaiveBayes.jl does that different again, givein tuples of class, logprob…)
In anycase some good old fashion NaiveBayes classification right here.
Right now support for julia 0.6 hasn’t been merged.
So you need to use the branch from PR #35
Do this via Pkg.checkout("NaiveBayes.jl", "julia-0.6-2").
Input:
import NaiveBayes: predict_logprobs, HybridNB, restructure_matrixOutput:
WARNING: Method definition (::Type{KernelDensity.InterpKDE{K, I} where I where K})(KernelDensity.UnivariateKDE{R} where R<:(Base.Range{T} where T)) in module KernelDensity at /home/wheel/oxinabox/.julia/v0.6/KernelDensity/src/interp.jl:15 overwritten in module NaiveBayes at /home/wheel/oxinabox/.julia/v0.6/NaiveBayes/src/hybrid.jl:133.Input:
fit(::Type{HybridNB}, features, labels) = fit(HybridNB(labels), features, labels)
function predict(model::HybridNB, features)
unnormed_logprobs = predict_logprobs(model, restructure_matrix(features), Dict{Symbol, Vector{Int}}())
ps = exp.(unnormed_logprobs)
(ps./sum(ps,1))[1,:]
endOutput:
predict (generic function with 9 methods)
Input:
evaluate(HybridNB)Output:
4.300476 seconds (1.46 M allocations: 76.450 MiB, 1.53% gc time)
NaiveBayes.HybridNB Train accuracy: 92.11%
NaiveBayes.HybridNB Test accuracy: 90.58%Bloby. Very blobby. I’m not really to sure why it acts this way. Of all theses classifiers Naive Bayes is the one I least know the internals of.
TensorFlow.jl
TensorFlow.jl is my baby: Ok, it is actually @malmaud’s baby. I guess I’m that cool uncle :-P that teach’s it how to make slingshots and things; or in this case do julia-style indexing and such.
Using a neural network to do classification is a bit like driving a tank to the shops. Sure it will get you there; and nothing can ever stop you getting there. But it’s not exactly fast. So maybe you stop and give up before reaching the store.
Since it is a general Neural Net framework (and a low level one at that), we have to build the classifier ourself.
Input:
using TensorFlow
struct TensorFlowClassifier
xs::Tensor{Float32}
ys::Tensor{Float32}
ys_targets::Tensor{Float32}
mean_loss::Tensor{Float32}
optimizer::Tensor # weirdly this is not type-stable, probably a bug I need to chase down. It *should* always be `Tensor{Any}`
sess::Session
endInput:
function TensorFlowClassifier(nfeatures, hidden_layers)
layer_sizes = [nfeatures; hidden_layers; 1]
sess = Session(Graph())
@tf begin
xs = placeholder(Float32, shape=[nfeatures, -1]) #Nfeatures, Nsamples (going to use julian ordering)
zs = xs
for ii in 2:length(layer_sizes)
size_below = layer_sizes[ii-1]
size_above = layer_sizes[ii]
W = get_variable("W_$ii", [size_above, size_below], Float32)
b = get_variable("b_$ii", [size_above, 1], Float32)
zs = nn.sigmoid(W*zs + b; name="zs_$ii")
end
ys = squeeze(zs, [1]) #drop first dimention
ys_targets = placeholder(Float32, shape=[-1])
loss = nn.sigmoid_cross_entropy_with_logits(;logits=ys, targets=ys_targets)
mean_loss = mean(loss)
optimizer = TensorFlow.train.minimize(train.AdamOptimizer(), loss)
end
run(sess, global_variables_initializer())
TensorFlowClassifier(xs,ys, ys_targets, mean_loss, optimizer, sess)
endOutput:
TensorFlowClassifier
Input:
function fit(::Type{TensorFlowClassifier}, features, labels; hidden_layers=[256, 256], kwargs...)
nfeatures = size(features, 1)
model = TensorFlowClassifier(nfeatures, hidden_layers)
fit!(model, features, labels; kwargs...)
end
function fit!(model::TensorFlowClassifier, features, labels; silent=true, max_epochs=100_000, conv_atol=0.005, conv_period=5)
conv_timer = conv_period
old_loss_o = Inf
for ii in 1:max_epochs
loss_o, _ = run(model.sess, (model.mean_loss, model.optimizer), Dict(model.xs=>features, model.ys_targets=>labels))
if ii%1_000==1
!silent && println(loss_o)
# Check for convergence
if loss_o < old_loss_o - conv_atol
conv_timer = conv_period
else
conv_timer-=1
if conv_timer < 1
break
end
end
end
old_loss_o = loss_o
end
model
endOutput:
fit! (generic function with 3 methods)
Input:
function predict(model::TensorFlowClassifier, features)
run(model.sess, model.ys, Dict(model.xs=>features))
endOutput:
predict (generic function with 10 methods)
Input:
evaluate(TensorFlowClassifier)Output:
2017-12-18 15:40:48.217612: W tensorflow/core/platform/cpu_feature_guard.cc:45] The TensorFlow library wasn't compiled to use SSE4.1 instructions, but these are available on your machine and could speed up CPU computations.
2017-12-18 15:40:48.217647: W tensorflow/core/platform/cpu_feature_guard.cc:45] The TensorFlow library wasn't compiled to use SSE4.2 instructions, but these are available on your machine and could speed up CPU computations.158.325735 seconds (11.66 M allocations: 938.312 MiB, 0.39% gc time)
TensorFlowClassifier Train accuracy: 93.99%
TensorFlowClassifier Test accuracy: 91.79%This is not a nice kinda problem for a neural network. The neural network kind of relies on luck to start out with something that it can gradient descent towards a good hidden representation. Having a wide hidden layer helps with that (also the hidden layer is outright required to learn somplex projections).
The big problem though is the highly unbalanced data. There probably is a gradient for the it to decend down towards getting those stars to show up. But it is really shallow.
If this were a NN blog post I would talk about and show how to fix it. But this isn’t.
We’re looking at many libraries and many methods. I will show another neural one, so the code can be compared.
Flux
Flux is the new hotness when it comes to julia neural network frameworks.

Julia has a fair few neural net frameworks. As well as the already mentioned TensorFlow.jl, we’ve got MXNet.jl (which, like TensorFlow.jl is a wrapper), we’ve got Knet.jl, Mocha.jl and probably a few others that I am forgetting. I don’t have time to go through and show how to make them all run. But I’ll do Flux.
Flux.jl has some really nice clean julia design; in ways that were not possible (do to language and ecosystem development) when the other pure julia options came out.
As with TensorFlow I have to define a binary classifier myself. Also flux doesn’t have the binary cross entropy function (Related: closed PR #96, so better define that.
This code is running against Flux PR #132, so you’ll need to checkout that branch.
Input:
using FluxInput:
function fit(::Type{Flux.Chain}, features, labels; hidden_layers=[256, 256], kwargs...)
nfeatures = size(features, 1)
layer_sizes = [nfeatures; hidden_layers; 1]
layers = map(2:length(layer_sizes)) do ii
size_below = layer_sizes[ii-1]
size_above = layer_sizes[ii]
Dense(size_below, size_above, σ)
end
model = Chain(layers..., transpose)
fit!(model, features, labels; kwargs...)
endOutput:
fit (generic function with 31 methods)
Input:
"""
iter_throttle(f, period; default_return=false)
Helper function to only run `f` every `period` times that it is called.
In rounds which it does not run, returns `default_return`
"""
function iter_throttle(f, period; default_return=false)
round = 0
function(args...)
if round==period
round=0
f(args...)
else
round+=1
default_return
end
end
endOutput:
iter_throttle
Input:
function fit!(model::Flux.Chain, features, labels; silent=true, max_epochs=100_000, conv_atol=0.005, conv_period=5)
function loss(x, y) # AFAIK Flux loss must *always* be a closure around the model (or the model be global)
ŷ = model(x)
-sum(log.(ifelse.(y, ŷ, 1-ŷ))) #Binary Cross-Entropy loss
end
old_loss_o = Inf
conv_timer = conv_period
function stop_cb(ii,loss_val)
loss_o = loss_val/length(labels) #real loss is the sum, but we want to use the mean
if loss_o < old_loss_o - conv_atol
conv_timer = conv_period
else
conv_timer-=1
if conv_timer < 1
return true
end
end
old_loss_o = loss_o
return false
end
log_cb(ii, loss_val) = println("at $ii loss: ", loss_val/length(labels))
opt = ADAM(params(model))
dataset = Base.Iterators.repeated((features, labels), max_epochs)
Flux.train!(loss, dataset, opt,
log_cb = Flux.throttle(silent ? (i,l)->(): log_cb, 10), # every 10 seconds
stopping_criteria = iter_throttle(stop_cb, 100), #Every 100 rounds
)
model
endOutput:
fit! (generic function with 4 methods)
Input:
function predict(model::Flux.Chain, features)
Flux.Tracker.data(model(features))
endOutput:
predict (generic function with 11 methods)
Note that I am running the stopping criteria check every 100 round, rather than every 1000 rounds as I was for TensorFlow. because it seems to me like running 1000 rounds in Flux takes like 10 minutes. So I’m willing to let terminate early, rather than continue to take its time getting those lost bits of accuracy out.
Flux really seems slow, but I’ld have to be a bit more careful with my tests to make sure of that. Particularly since for both Flux and TensorFlow we are not running for a fixed number of rounds, but until convergence is met. Might be a future blog post, doing a tight performance test between NN frameworks.
Input:
evaluate(Flux.Chain)Output:
552.606535 seconds (660.42 k allocations: 131.764 GiB, 59.47% gc time)
Flux.Chain Train accuracy: 93.87%
Flux.Chain Test accuracy: 92.05%Wow, Flux allocated and deallocated a lot of memory.
TensorFlow.jl may have done the same, but you can’t see it so much, because it can hide behind the ccall to the backend.
I suspect it didn’t though.
Note that the networks for Flux and TensorFlow were identical, in terms of activation functions, loss functions and layer-sizes. The difference comes in the intitalisation and training hyper-paramaters. It ended up marginally outperforming the Tesnorflow network, not because of structual differences, but because of these optimisation hyper-paramater differences.
I suspect the blurryness with factional probabilities is because flux wasn’t given enough time to fully converge.
I particularly like the downwards streak from the union jack to the commonwealth start (ends as x=100).
That is very cool.
This really highlights that neural networks are working with approximating continous smooth functions.
It is harder for it to learn that disconnected region, since the function would have to go to one, then flip back down to zero.
Given enough hidden layer neurons and time, it probably would have fitted that, and disconnected the commonwealth start from the union jack.
Conclusion
So there are a lot of ways to do binary classification with julia packages. We saw 7, and there are a few I didn’t get to include. I’m particular sad to miss GLMNet.jl. I honestly couldn’t workout how to use it. I hear it is great system.
We looked at 4 families of methods. Support vector machines, with LIBLINEAR.jl and LIBSVM.jl. Decision Trees and similar, with DecisionTrees.jl and XGBoost.jl. Naive Bayes, with NaiveBayes.jl. And finally neural networks, with TensorFlow.jl, and Flux.jl
I guess the take away might be don’t use neural networks for simple binary classification. They are slow, they don’t work well, they have too many hyper-parameters, and they don’t come with nice interface. However I don not think that it nessicarily completely valid.
With regards to the slowness, I think they’ld be faster if one were willing to tweak the learning-rate up a bit. The right set of layer-size hyper-parameters, any maybe some tricks to rebalance the classes, and normalize the input I think would improve performane immensely. Finally as to the nice interface, that is not so much a property of the neural network so much as in my choice of libraries. As a ML researcher I tend to be most interested in the low level libraries that give me full control. But other interfaces can hide a lot of the code I wrote above – as a random example the Scikit Learn MLPClassifier.
Of course this is only simple binary classification. By simple I mean there are 2 input features, being the pixel coordinates. Image classification would have thousand or millions of input features (depending on resolution). For image classification convolutional neural networks are king. Though don’t forget there is the option of using image embeddings, i.e. CNN features (e.g. from a pretrained network like Inception, or another ImageNet network), as the input another classifier. I know people do this with SVM’s all the time.
I do think a take-way is that neural networks probably shouldn’t be your first choice for simple binarly classification. Looking at how well everything else performs, it is almost always going to be worth checking the others out. Their training time is so short, it is worth checking them out, as its not going to cost much time to evaluate them.